Reshaping data with tidyr
One of the key task in data preparation is to organize thee dataset in a way that makes analysis and plottng easier. In practice, the data is often not stored like that and the data comes to us with repeated observations included on a single row. This is often done as a memory saving technique or because there is some structure in the data that makes the ‘wide’ format attractive. As a result, we need a way to convert data from wide1 to long2 and vice-versa [@mathew].
Structuring data frames to have the desired shape can be the most daunting part of statistical analysis, visualisation, and modeling. Several studies reported that 80% of data analysis is spent on the cleaning and preparing data. Tidy in this context means organize the data in a presentable and consistent format that facilitate data analysis and visualization. When you are doing data preparation in R for analysis or plottng, the first thing you do is a throughly mental thought on the desired structure of that data frame. You need to determine what each row and column will represent, so that you can consistently and clearly manipulate that data (e.g., you know what you will be selecting and what you will be filtering).
There are basically three principles that we can follow to make a tidy dataset. First each variable must have its own a column, second each observation must have its own row, and finally, each cell must have its own value. The tidyr package is used to structure and work with data fames that follow three principles of tidy data. There are three advantages of using tidy data in R. First, having a consistent, uniform data structure is very important. Popular packages like dplyr [ @dplyr], ggplot2 [@ggplot], and all the other packages in the tidyverse [@tidyverse] and their extensions like sf [@sf], metR [@metr], ggspatial [@ggspatial], ggrepel [@ggrepel] etc are designed to work with tidy data [@tidyr]. So consistent of tidy data ensure efficient processing, analysis and plotting of data. Third, placing variables into columns, facilities easy vectorization in R.
Unfortunate, Many datasets that you receove are untidy and will require some work on your end. There are several reasons why a dataset messy. Often times the people who created the dataset aren’t familiar with the principles of tidy data. Another common reason that most datasets are messy is that data is often organized to facilitate something other than analysis. Data entry is perhaps the most common of the reasons that fall into this category. To make data entry as easy as possible, people will often arrange data in ways that aren’t tidy. So, many datasets require some sort of tidying before you can begin your analysis.
As @r4d put it tidy data means that yo can plot or summarize the data efficiently. In othet words, it comes down to which data is represented as columns in a data frame and which is not.In principle, this means that there is column in the data frame that you can work with for the analysis you want to do. For example, if I want to look at the ctd dataset and see how the fluorescence varies among station in the upper water surface, we simply plot a boxplot of the station column against the fluorescence column shown in figure 1
require(tidyverse)
require(magrittr)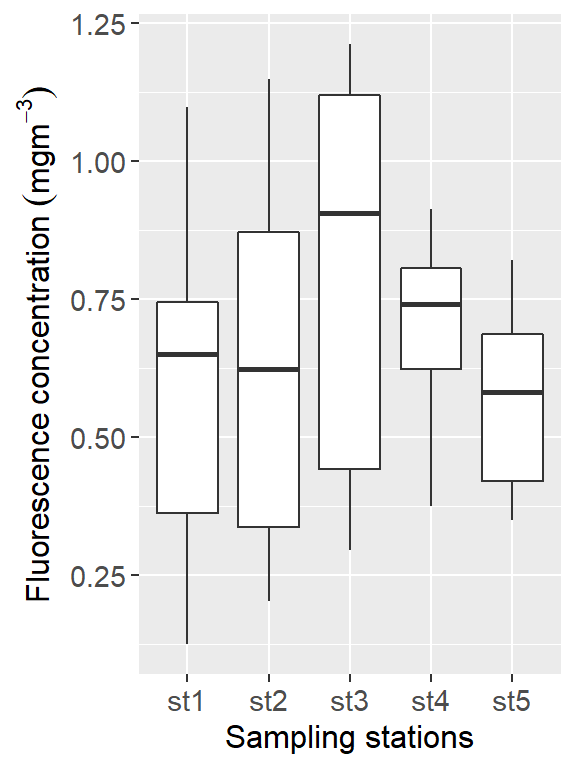
Figure 1: Fluorescence variation against stations
This works because I have a column for the x-axis and another for the y-axis. But what happens if I want to plot the different measurements of the irises to see how those are? Each measurement is a separate column. They are Petal.Length, Petal.Width, and so on.Now I have a bit of a problem because the different measurements are in different columns in my data frame. I cannot easily map them to an x-axis and a y-axis.The tidyr package addresses that. It contains functions for both mapping columns to values—, widely recognised as long format and for mapping back from values to columns—wide format.
We are going to look for the function that are regularly used to tidy the data frames. These inlude:
- Gathering
- Spreading
- Separating
- Uniting
Gather—from wide to long format.
Look at example of dataset. It has one common problem that the column names are not variables but rather values of a variable. In the table 1, the columns are actually values of the variable pressure. Each row in the existing table actually represents five observations from each station.
| station | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| st1 | 0.599 | 0.678 | 0.729 | 0.693 | 0.752 | 1.098 | 0.857 | 0.481 | 0.313 | 0.221 | 0.138 |
| st2 | 0.631 | 0.733 | 0.992 | 1.114 | 0.988 | 0.715 | 0.496 | 0.524 | 0.280 | 0.277 | 0.225 |
| st3 | 0.980 | 0.934 | 1.149 | 1.200 | 1.187 | 1.035 | 0.854 | 0.530 | 0.437 | 0.347 | 0.324 |
| st4 | NA | 0.623 | 0.603 | 0.742 | 0.724 | 0.799 | 0.914 | 0.819 | 0.801 | 0.692 | 0.444 |
| st5 | NA | 0.350 | 0.415 | 0.421 | 0.566 | 0.593 | 0.591 | 0.634 | 0.751 | 0.774 | 0.575 |
The tidyr package can be used to gather these existing columns into a new variable. In this case, we need to create a new column called pressure and then gather the existing values in the these variable columns into the new pressure variable
ctd.long = ctd.wide %>%
gather (key = "depth", value = "fluorescence ", 2:12)
ctd.long# A tibble: 55 x 3
station depth `fluorescence `
<chr> <chr> <dbl>
1 st1 10 0.599
2 st2 10 0.631
3 st3 10 0.98
4 st4 10 NA
5 st5 10 NA
6 st1 20 0.678
7 st2 20 0.733
8 st3 20 0.934
9 st4 20 0.623
10 st5 20 0.350
# ... with 45 more rowsAs you can see from the chunk above, there are three arguments required in the gather() function. First is the key, which takes the variable names. Second, the value—the name of the variable whose values are spread over the cells. Finnaly, then you specify the set of columns that hold the values and not the variable names
spread() —from long to wide format
A second tidy tool is spread(), which does the opposite of gather() function. It is used to convert a long format data frame to wide format. What this function does is to spread observation across multiple rows.
ctd.long %>%
spread(key = "depth", value = `fluorescence `)# A tibble: 5 x 12
station `10` `100` `110` `20` `30` `40` `50` `60` `70` `80` `90`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 st1 0.599 0.221 0.138 0.678 0.729 0.693 0.752 1.10 0.857 0.481 0.313
2 st2 0.631 0.277 0.225 0.733 0.992 1.11 0.988 0.715 0.496 0.524 0.280
3 st3 0.98 0.347 0.324 0.934 1.15 1.20 1.19 1.03 0.854 0.530 0.437
4 st4 NA 0.692 0.444 0.623 0.603 0.742 0.724 0.799 0.914 0.819 0.801
5 st5 NA 0.774 0.575 0.350 0.415 0.421 0.566 0.592 0.591 0.634 0.751The spread() function takes two arguments: the column that contains variable names, known as the key and
a column that contains values from multiple variables – the value.
separate()
Another common in tidyr package is a separate ()function, which split the variable into two or more variables. For example, the dataset below has a date column that actually contains the date and time variables separated by a space.
ctd# A tibble: 115 x 12
station time lon lat pressure temperature salinity oxygen
<chr> <dttm> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 st1 2004-08-18 15:27:46 40.6 -10.5 5 25.2 33.9 3.93
2 st1 2004-08-18 15:27:46 40.6 -10.5 10 25.1 34.9 4.49
3 st1 2004-08-18 15:27:46 40.6 -10.5 15 25.1 34.9 4.50
4 st1 2004-08-18 15:27:46 40.6 -10.5 20 25.0 34.9 4.51
5 st1 2004-08-18 15:27:46 40.6 -10.5 25 24.9 34.9 4.51
6 st1 2004-08-18 15:27:46 40.6 -10.5 30 24.9 34.9 4.50
7 st1 2004-08-18 15:27:46 40.6 -10.5 35 24.9 34.9 4.49
8 st1 2004-08-18 15:27:46 40.6 -10.5 40 24.9 34.9 4.48
9 st1 2004-08-18 15:27:46 40.6 -10.5 45 24.8 34.9 4.46
10 st1 2004-08-18 15:27:46 40.6 -10.5 50 24.6 34.9 4.44
# ... with 105 more rows, and 4 more variables: fluorescence <dbl>, spar <dbl>,
# par <dbl>, density <dbl>We use the separate() function splits the datetime column into two variables: date and time
ctd %>%
separate(col = time, into = c("Date", "Time"), sep = " ")# A tibble: 115 x 13
station Date Time lon lat pressure temperature salinity oxygen
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 st1 2004-08-18 15:27:46 40.6 -10.5 5 25.2 33.9 3.93
2 st1 2004-08-18 15:27:46 40.6 -10.5 10 25.1 34.9 4.49
3 st1 2004-08-18 15:27:46 40.6 -10.5 15 25.1 34.9 4.50
4 st1 2004-08-18 15:27:46 40.6 -10.5 20 25.0 34.9 4.51
5 st1 2004-08-18 15:27:46 40.6 -10.5 25 24.9 34.9 4.51
6 st1 2004-08-18 15:27:46 40.6 -10.5 30 24.9 34.9 4.50
7 st1 2004-08-18 15:27:46 40.6 -10.5 35 24.9 34.9 4.49
8 st1 2004-08-18 15:27:46 40.6 -10.5 40 24.9 34.9 4.48
9 st1 2004-08-18 15:27:46 40.6 -10.5 45 24.8 34.9 4.46
10 st1 2004-08-18 15:27:46 40.6 -10.5 50 24.6 34.9 4.44
# ... with 105 more rows, and 4 more variables: fluorescence <dbl>, spar <dbl>,
# par <dbl>, density <dbl>The separate() function accepts arguments for the name of the variable to separate. You also need to specify the names of the variable to separate into, and an optional separator.
##unite ()
The unite()function is the exact opposite of separate() in that it combines multiple columns into a single column. While not used nearly as often as separate() , there may be times when you need the functionality provided by unite(). For example, we can combine the variable Date and Time to form siku.muda, and separate them with :-: symbol between the two variables.
ctd.un %>% unite(col = "siku_muda", c("Date", "Time"), sep = ":-:")# A tibble: 115 x 12
station siku_muda lon lat pressure temperature salinity oxygen
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 st1 2004-08-18:-:15:27:~ 40.6 -10.5 5 25.2 33.9 3.93
2 st1 2004-08-18:-:15:27:~ 40.6 -10.5 10 25.1 34.9 4.49
3 st1 2004-08-18:-:15:27:~ 40.6 -10.5 15 25.1 34.9 4.50
4 st1 2004-08-18:-:15:27:~ 40.6 -10.5 20 25.0 34.9 4.51
5 st1 2004-08-18:-:15:27:~ 40.6 -10.5 25 24.9 34.9 4.51
6 st1 2004-08-18:-:15:27:~ 40.6 -10.5 30 24.9 34.9 4.50
7 st1 2004-08-18:-:15:27:~ 40.6 -10.5 35 24.9 34.9 4.49
8 st1 2004-08-18:-:15:27:~ 40.6 -10.5 40 24.9 34.9 4.48
9 st1 2004-08-18:-:15:27:~ 40.6 -10.5 45 24.8 34.9 4.46
10 st1 2004-08-18:-:15:27:~ 40.6 -10.5 50 24.6 34.9 4.44
# ... with 105 more rows, and 4 more variables: fluorescence <dbl>, spar <dbl>,
# par <dbl>, density <dbl>
